IUST-Networks
Sample projects and assignments in dynamic complex networks
Introduction
This repository contains my assignments and projects submitted as a requirement of IUST Dynamic Complex Network course.
Hafez (حافظ) Wordnet
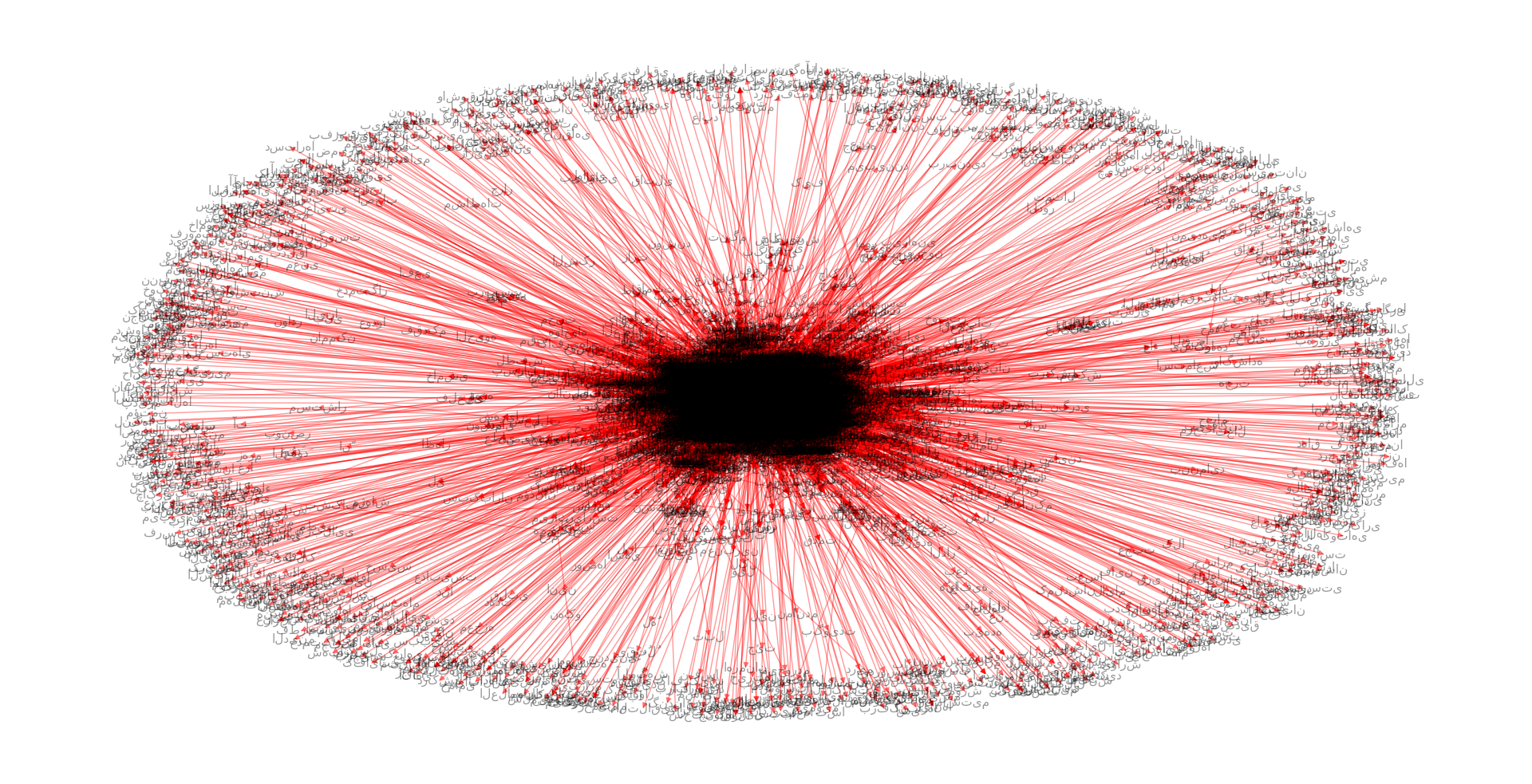
We extracted a word network from Hafez poems. A naive version of this network is presented here. Each word is connected to the next word in a same poetry. A subgraph cosist of first 100 nodes is shown in below figure.
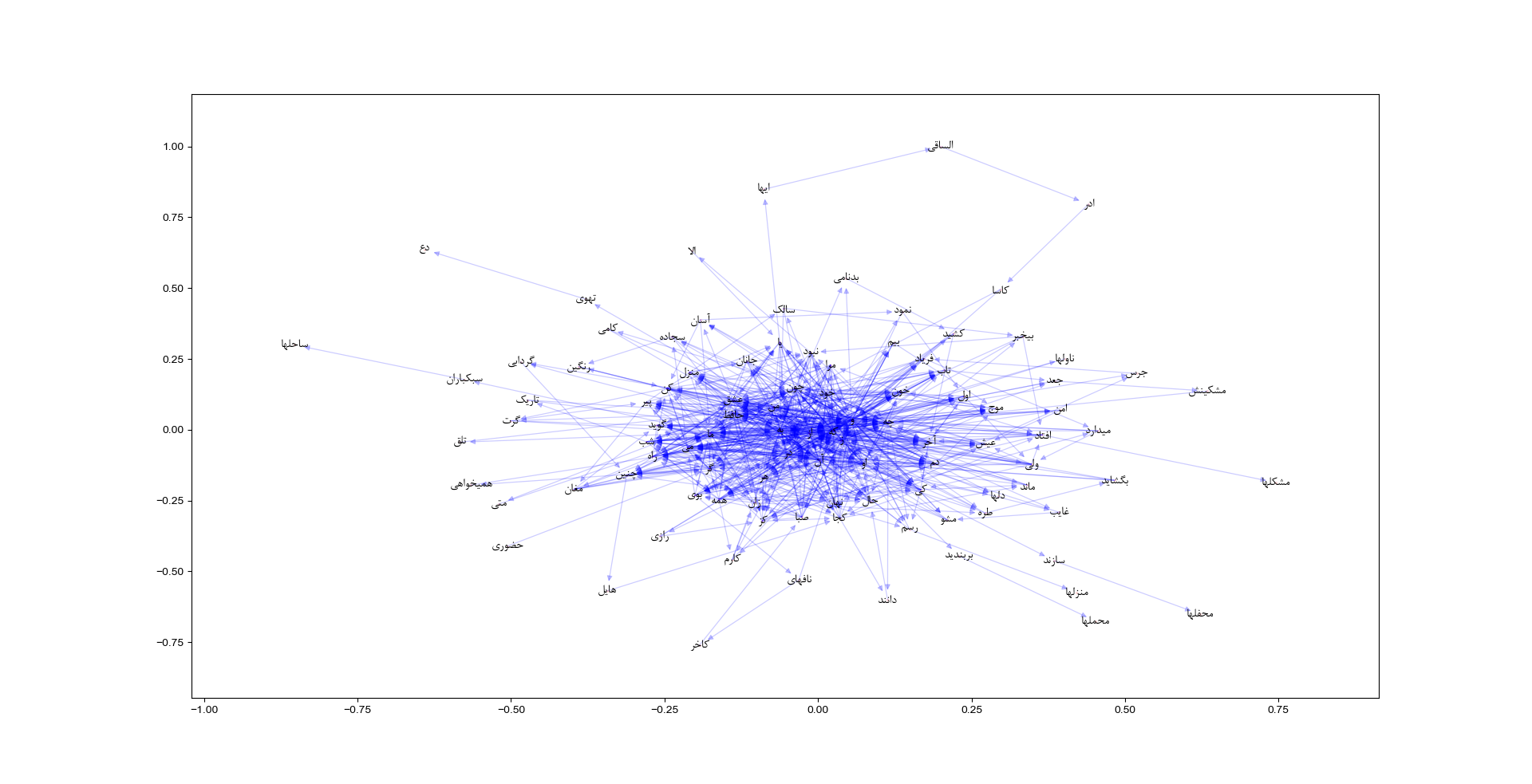
The above network is based on the preamble poetry in Divan-e-Hafez:

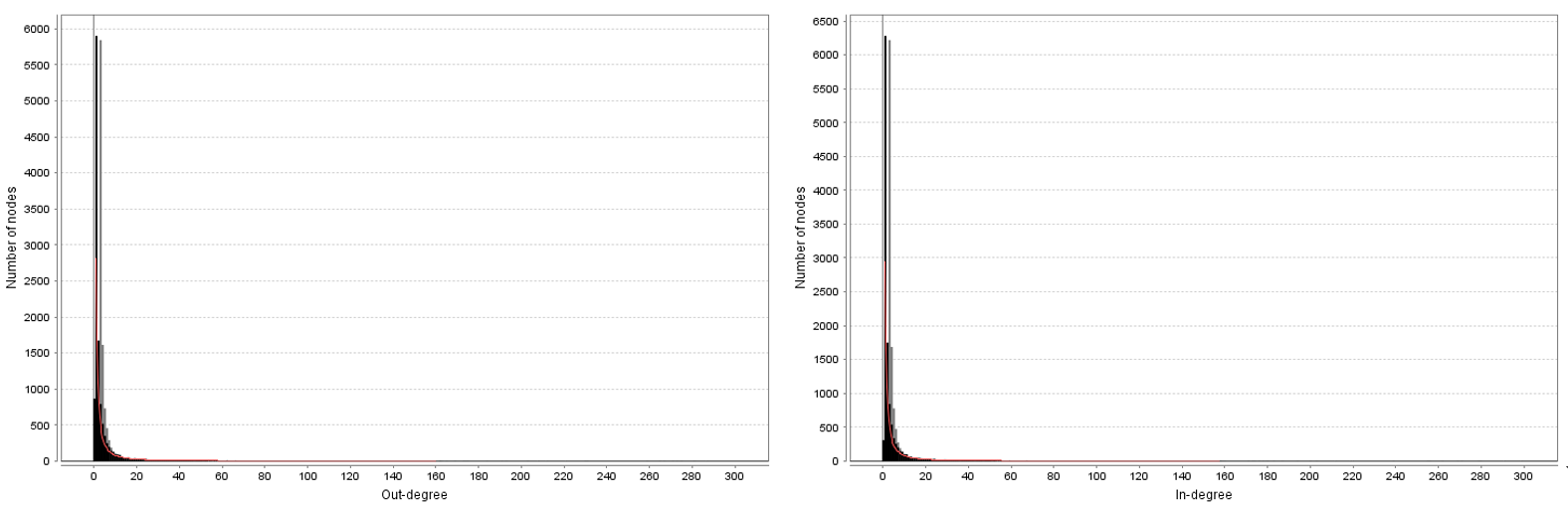
Degree Distrubution
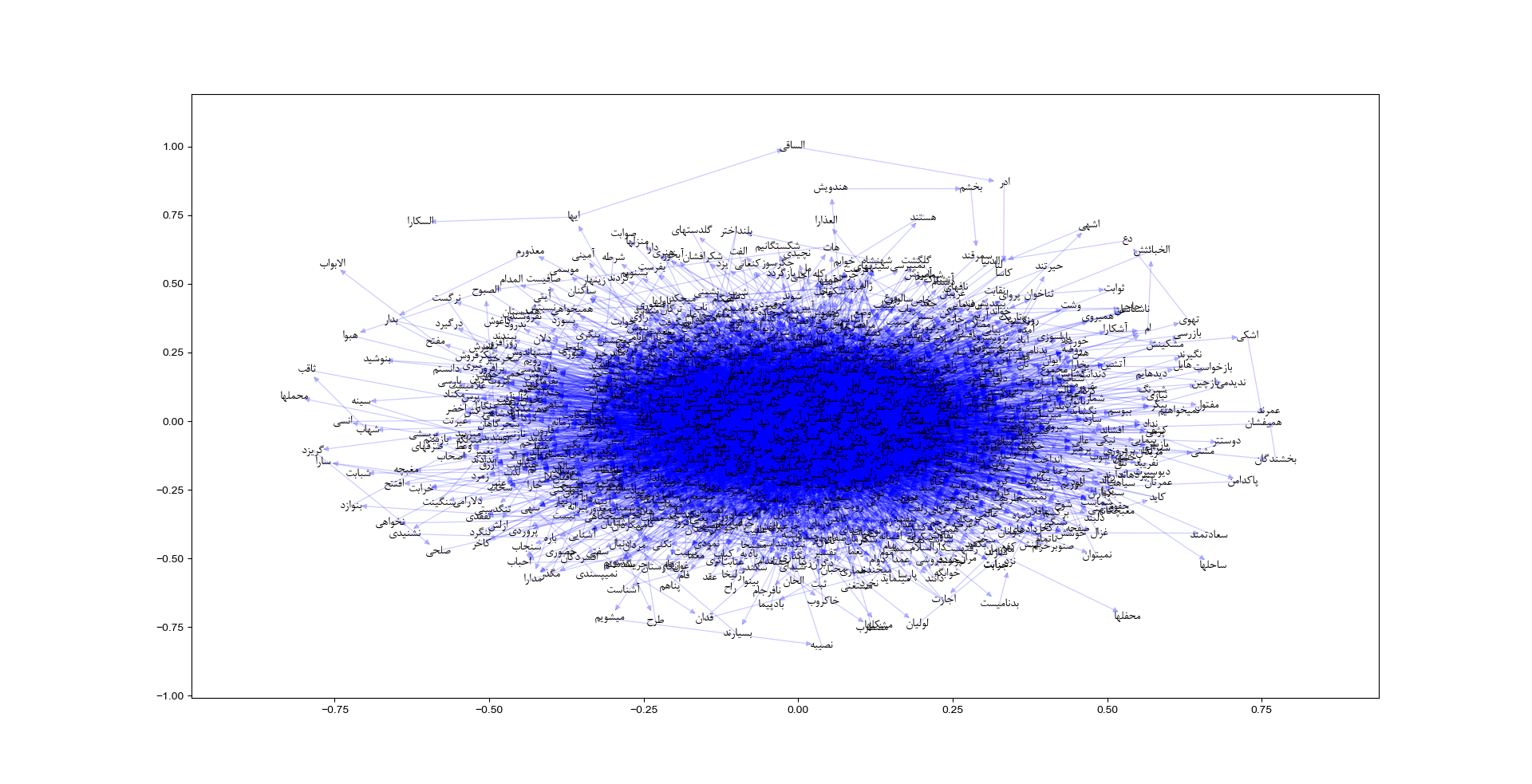
Hafez Poems (Subgraph with 1000 Nodes)
Network Analysis for Hafez Wordnet
To be complete . . .
Assignment Details
Assignment 3
1) Collect some networks 2) Compute some metrics of these networks 3) Analysis and discuss the results
Assignment 4
Consider the three network datasets of assignment #3
1) Fit E-R, W-S, B-A, and X models to these networks 2) Select (find) X as the best fitting model 3) Generate artificial graphs similar to that networks * Specify the suitable generative parameters 4) Compute some macro-level metrics * Degree distribution, avg clustering coefficient, … 5) Compare the features of the real and artificial graph counterparts 6) Analyze the results (comparison)
Assignment 5
Consider the three network datasets of previous assignments 1) Find communities in each network 2) Report “modularity” of the communities.
Assignment 6
Simulate two epidemic models on two network models 1) Four simulation scenarios * E.g., SIRS epidemic model on BA network model
The importance of epidemy models is obvious in cases such as COVID-19 pandemic. Here we are going to perform some epidemics simulation on different graph models and depicting the result.
SIR on Barabasi-Albert graph
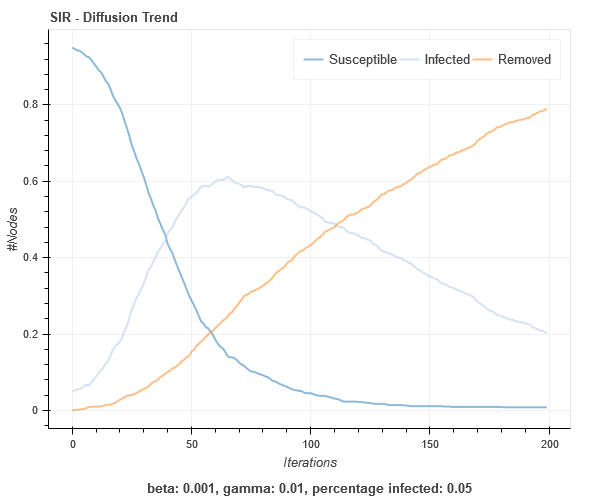
SIS on Barabasi-Albert graph
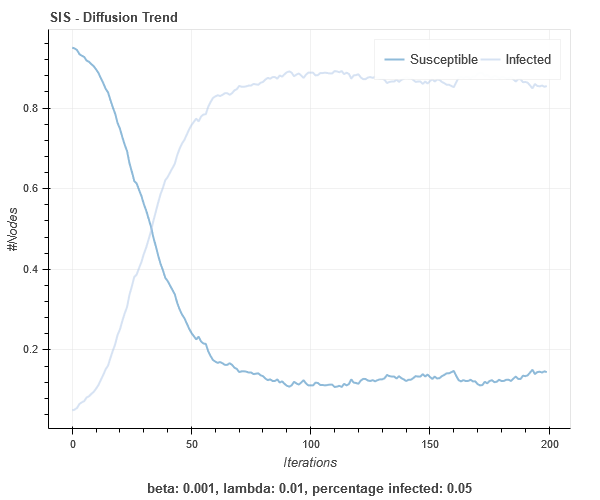
References
[1] J. Leskovec, J. Kleinberg and C. Faloutsos. Graphs over Time: Densification Laws, Shrinking Diameters and Possible Explanations. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD), 2005.
[2] J. Gehrke, P. Ginsparg, J. M. Kleinberg. Overview of the 2003 KDD Cup. SIGKDD Explorations 5(2): 149-151, 2003.
[3] J. Leskovec, J. Kleinberg and C. Faloutsos. Graph Evolution: Densification and Shrinking Diameters. ACM Transactions on Knowledge Discovery from Data (ACM TKDD), 1(1), 2007.
[4] Cytoscape, “Cytoscape.” [Online]. Available: https://cytoscape.org/. [Accessed: 25-Apr-2019].
[5] NetworkX, “NetworkX.” [Online]. Available: https://networkx.github.io/. [Accessed: 26-Apr-2019].
[6] M. E. J Newman ‘Networks: An Introduction’, page 224, Oxford University Press 2011.
[7] Clauset, A., Newman, M. E., & Moore, C. “Finding community structure in very large networks.” Physical Review E 70(6), 2004.
[8] U. of Graz, “A (partially) interactive introduction to systems sciences.” [Online]. Available: http://systems-sciences.uni-graz.at/etextbook/networks/sirnetwork.html. [Accessed: 02-Jun-2019].
[9] G. Rossetti, L. Milli, S. Rinzivillo, A. Sirbu, F. Giannotti, and D. Pedreschi, “NDlib: a python library to model and analyze diffusion processes over complex networks,” CoRR, vol. abs/1801.0, 2018.
[10] NetworkX, “NetworkX.” [Online]. Available: https://networkx.github.io/. [Accessed: 26-Apr-2019].
[11] J. Banks and J. S. Carson, “Introduction to discrete-event simulation,” 2003, pp. 17–23.